A friend of mine pointed out that I had skipped a few steps in the solution I posted yesterday for Euler #3 – first, I didn’t actually go through the process of finding the primes, and second, I didn’t try to figure out how many prime numbers would be necessary for the list I used to find the largest prime factor of 600,851,475,143. To rectify these issues, I wrote another program that can provide the general solution for any integer larger than 1:
|
1 2 3 4 5 6 7 8 9 10 11 12 13 |
prime <- 2 primes <-c() remaining.factor <- 600851475143 while(prime != remaining.factor){ while(remaining.factor %% prime ==0){ remaining.factor <- remaining.factor/prime } primes <- append(primes,prime) while(length(which(prime %% primes ==0)>0)){ prime <- prime + 1 } } remaining.factor |
All you have to do is replace 600,851,475,143 with a number of your choice, and the script above will find the largest prime factor for that number, given enough time and memory. I was actually somewhat lucky that the answer ended up being 6857. Had it been larger, the program might have taken much longer to execute (possibly impractically long if the factor happened to be big enough).
Eratosthenes’ Sieve
Now that I have that out of the way, I would like to demonstrate a method for quickly generating prime numbers, called Eratosthenes’ Sieve. You can embed this algorithm into the solution of any Project Euler problem that calls for a large list of prime numbers under 10 million (after 10 million, the algorithm takes a long time to execute, and other sieves may be a better choice). While it’s possible to generate prime numbers on the fly for Euler problems, I still prefer to use lists to solve them. Below is the algorithm for Eratosthenes’ Sieve that I’ve written in R:
|
1 2 3 4 5 6 7 8 9 |
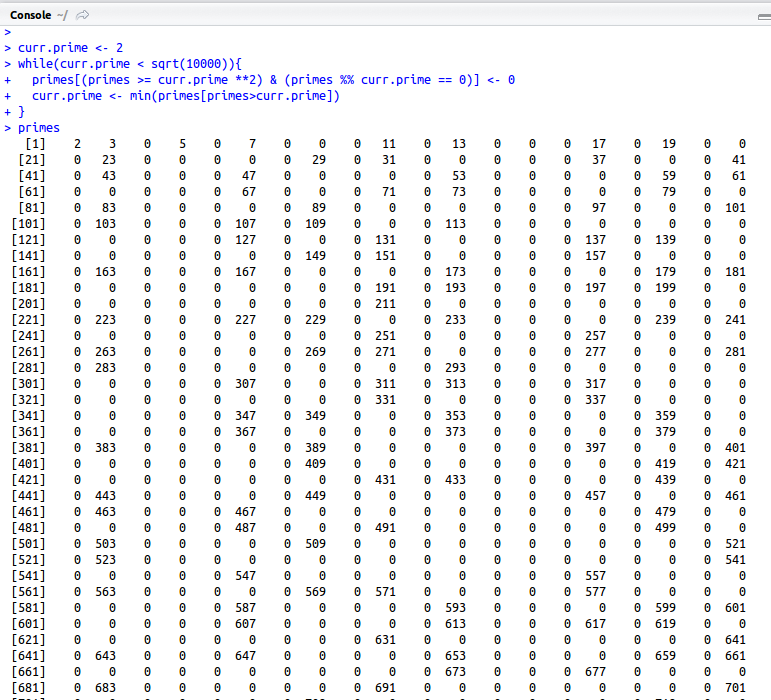
primes <- 2:10000000 curr.prime <- 2 while(curr.prime < sqrt(10000000)){ primes[(primes >= curr.prime **2) & (primes %% curr.prime == 0)] <- 0 curr.prime <- min(primes[primes>curr.prime]) } primes <- primes[primes!=0] |
The script above will generate a list of all the primes below 10,000,000. The algorithm starts with a list of integers from 2 to 10,000,000. Starting with 2, the smallest prime, you remove all the multiples of 2 from the list. Then you move on to the next prime, 3, and remove all the multiples of 3 from the list. The process continues until the only numbers left in the list are prime numbers.
As you can see, all the composite numbers are now marked as zeros. The final line of the script removes these zeros and gives you a list of just the prime numbers.